Draw a barplot of read totals
plotReadTotals(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'ANY'
plotReadTotals(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'FastqcDataList'
plotReadTotals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
duplicated = TRUE,
bars = c("stacked", "adjacent"),
vertBars = TRUE,
divBy = 1,
barCols = c("red", "blue"),
expand.y = c(0, 0.02),
plotlyLegend = FALSE,
...
)
# S4 method for class 'FastpDataList'
plotReadTotals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
adjPaired = TRUE,
divBy = 1e+06,
scaleFill = NULL,
labMin = 0.05,
status = TRUE,
labelVJ = 0.5,
labelFill = "white",
plotTheme = theme_get(),
vertBars = FALSE,
plotlyLegend = FALSE,
expand.y = c(0, 0.05),
...
)Arguments
- x
Can be a
FastqcData,FastqcDataListor file paths- usePlotly
logicalDefaultFALSEwill render using ggplot. IfTRUEplot will be rendered with plotly- labels
An optional named vector of labels for the file names. All filenames must be present in the names.
- pattern
Regex used to trim the end of filenames
- ...
Used to pass additional attributes to theme()
- duplicated
logical(1). Include deduplicated read total estimates to plot charts
- bars
If
duplicated = TRUE, show unique and deduplicated reads as "stacked" or "adjacent".- vertBars
logical(1) Show bars as vertical or horizontal
- divBy
Scale read totals by this value. The default shows the y-axis in millions for FastpDataList objects, but does not scale FastQC objects, for the sake of backwards compatability
- barCols
Colours for duplicated and unique reads.
- expand.y
Passed to
ggplot2::expansionfor the axis showing totals- plotlyLegend
logical(1) Show legend on interactive plots
- adjPaired
Scale read totals by 0.5 when paired
- scaleFill
ScaleDiscrete function to be applied to the plot
- labMin
Only show labels for filtering categories higher than this values as a proportion of reads. Set to any number > 1 to turn off labels
- status
logical(1) Include read status in the plot
- labelVJ
Relative vertical position to labels within each bar.
- labelFill
Passed to geom_label
- plotTheme
theme to be added to the plot
Value
Returns a ggplot or plotly object
Details
Draw a barplot of read totals using the standard ggplot2 syntax.
The raw data from readTotals() can otherwise be used to manually
create a plot.
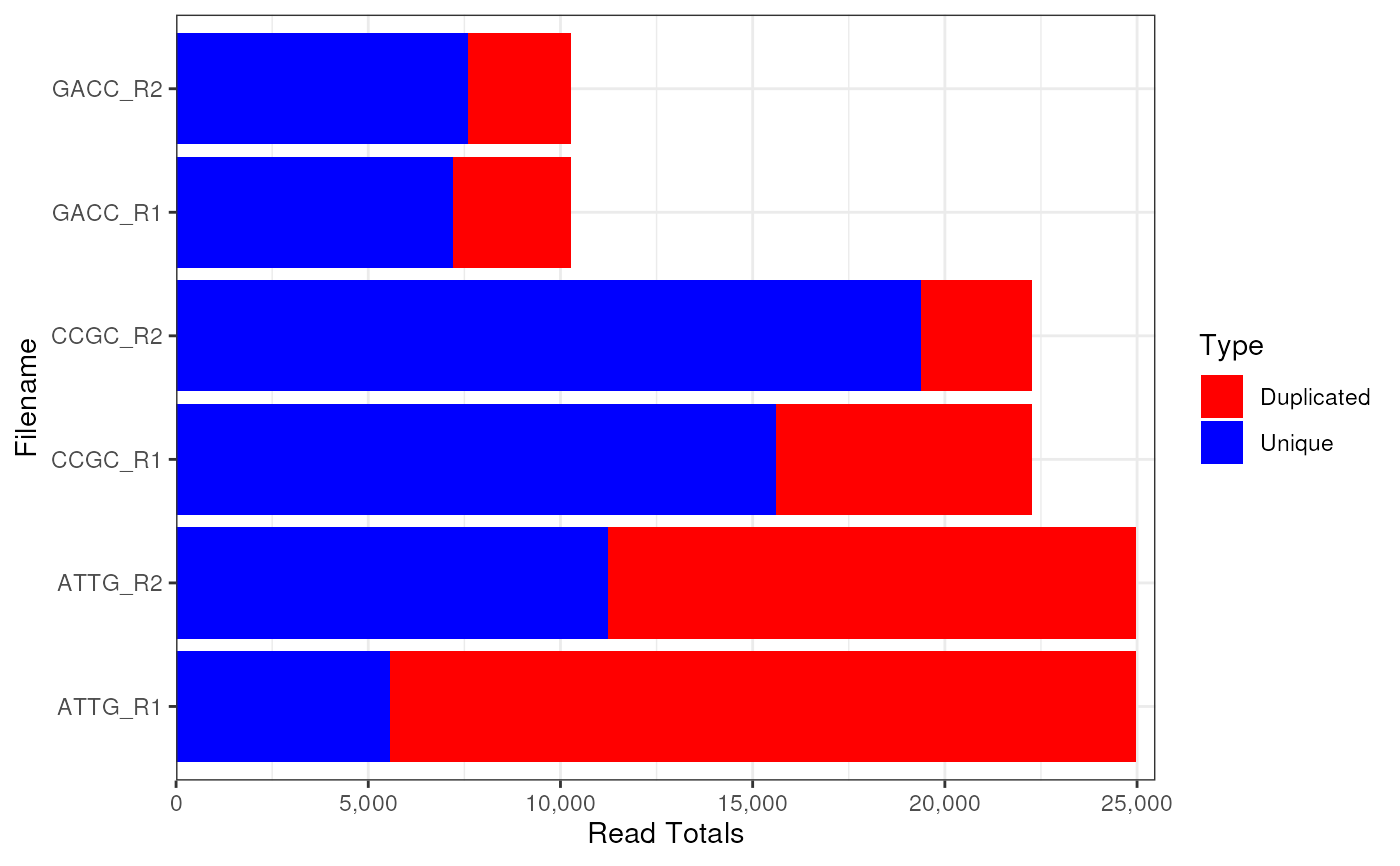
Duplication levels are based on the value shown on FASTQC reports at the
top of the DeDuplicatedTotals plot, which is known to be inaccurate.
As it still gives a good guide as to sequence diversity it is included as
the default. This can be turned off by setting duplicated = FALSE.
For FastpDataList objects, duplication statistics are not part of the default module containing ReadTotals. However, the status of reads and the reason for being retained or filtered is, and as such these are shown instead of duplication statistics.
Examples
# Get the files included with the package
packageDir <- system.file("extdata", package = "ngsReports")
fl <- list.files(packageDir, pattern = "fastqc.zip", full.names = TRUE)
# Load the FASTQC data as a FastqcDataList object
fdl <- FastqcDataList(fl)
# Plot the Read Totals showing estimated duplicates
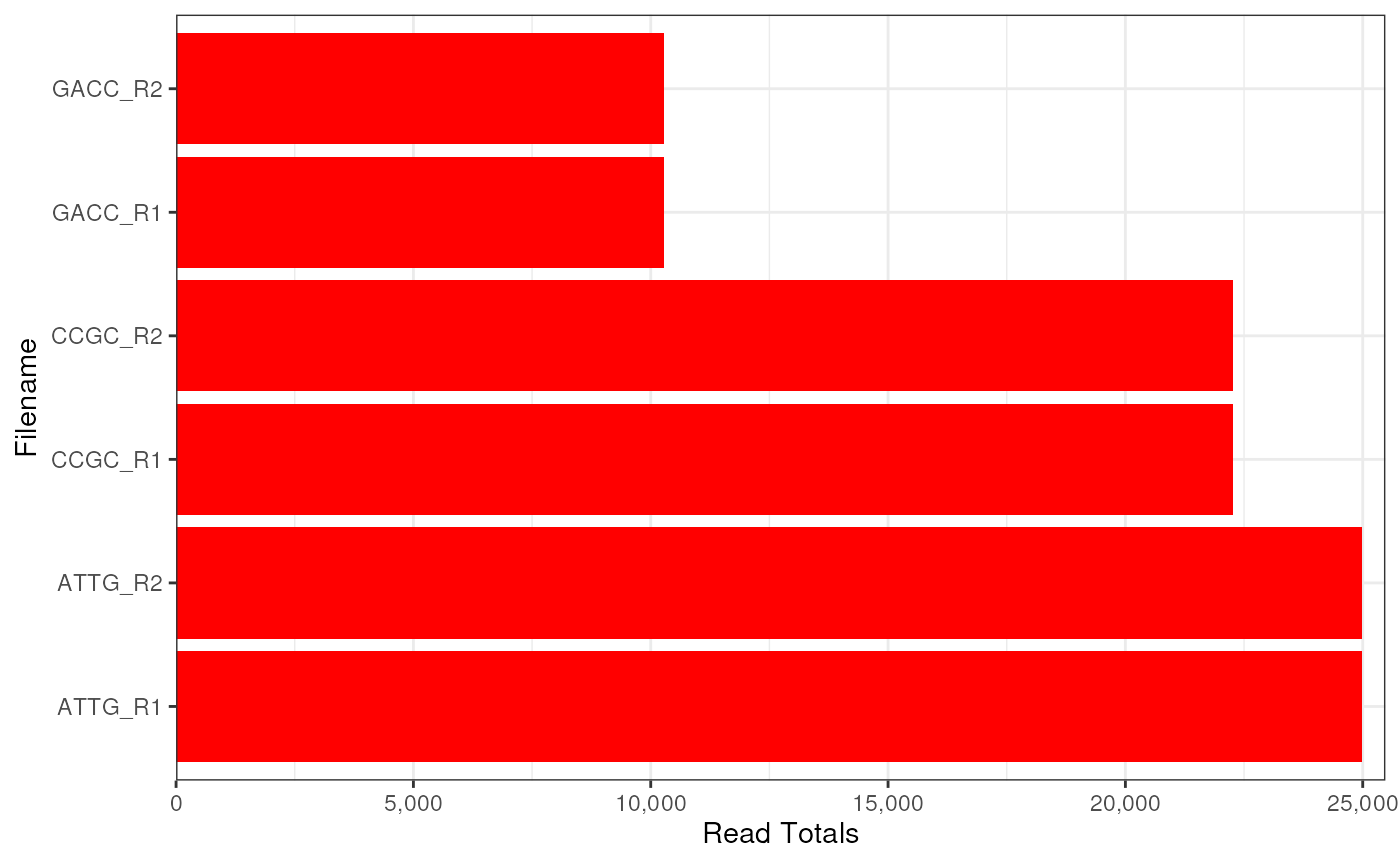
plotReadTotals(fdl)
 # Plot the Read Totals without estimated duplicates
plotReadTotals(fdl, duplicated = FALSE)
# Plot the Read Totals without estimated duplicates
plotReadTotals(fdl, duplicated = FALSE)