Plot Overrepresented Kmers
plotKmers(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'ANY'
plotKmers(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'FastqcData'
plotKmers(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
n = 6,
linewidth = 0.5,
plotlyLegend = FALSE,
scaleColour = NULL,
pal = c("red", "blue", "green", "black", "magenta", "yellow"),
...
)
# S4 method for class 'FastqcDataList'
plotKmers(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
cluster = FALSE,
dendrogram = FALSE,
pwfCols,
showPwf = TRUE,
scaleFill = NULL,
heatCol = hcl.colors(50, "inferno"),
heat_w = 8,
...
)
# S4 method for class 'FastpData'
plotKmers(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
module = c("Before_filtering", "After_filtering"),
reads = c("read1", "read2"),
readsBy = c("facet", "mean", "diff"),
trans = "log2",
scaleFill = NULL,
plotTheme = theme_get(),
plotlyLegend = FALSE,
...
)Arguments
- x
Can be a
FastqcData,FastqcDataListor file paths- usePlotly
logicalDefaultFALSEwill render using ggplot. IfTRUEplot will be rendered with plotly- labels
An optional named vector of labels for the file names. All filenames must be present in the names.
- pattern
regex to drop from the end of filenames
- ...
Used to pass parameters to theme for FastqcData objects and to geoms for FastpData objects
- n
numeric. The number of Kmers to show.- linewidth
Passed to
geom_line()- plotlyLegend
Show legend for interactive plots
- pal
The colour palette. If the vector supplied is less than n,
grDevices::colorRampPalette()will be used- cluster
logicaldefaultFALSE. If set toTRUE, fastqc data will be clustered using hierarchical clustering- dendrogram
logicalredundant ifclusterisFALSEif bothclusteranddendrogramare specified asTRUEthen the dendrogram will be displayed.- pwfCols
Object of class
PwfCols()to give colours for pass, warning, and fail values in the plot- showPwf
Show the PASS/WARN/FAIL status
- scaleFill, scaleColour
ggplot2 scales to be used for colour palettes
- heatCol
Colour palette used for the heatmap. Default is
infernofrom the viridis set of palettes- heat_w
Relative width of any heatmap plot components
- module
The module to obtain data from when using a FastpData object
- reads
Either read1 or read2. Only used when using a FastpData object
- readsBy
Strategy for visualising both read1 and read2. Can be set to show each set of reads by facet, or within the same plot taking the mean of the enrichment above mean, or the difference in the enrichment above mean
- trans
Function for transforming the count/mean ratio. Set as NULL to use the ratio without transformation
- plotTheme
theme object
Value
A standard ggplot2 object or an interactive plotly object
Details
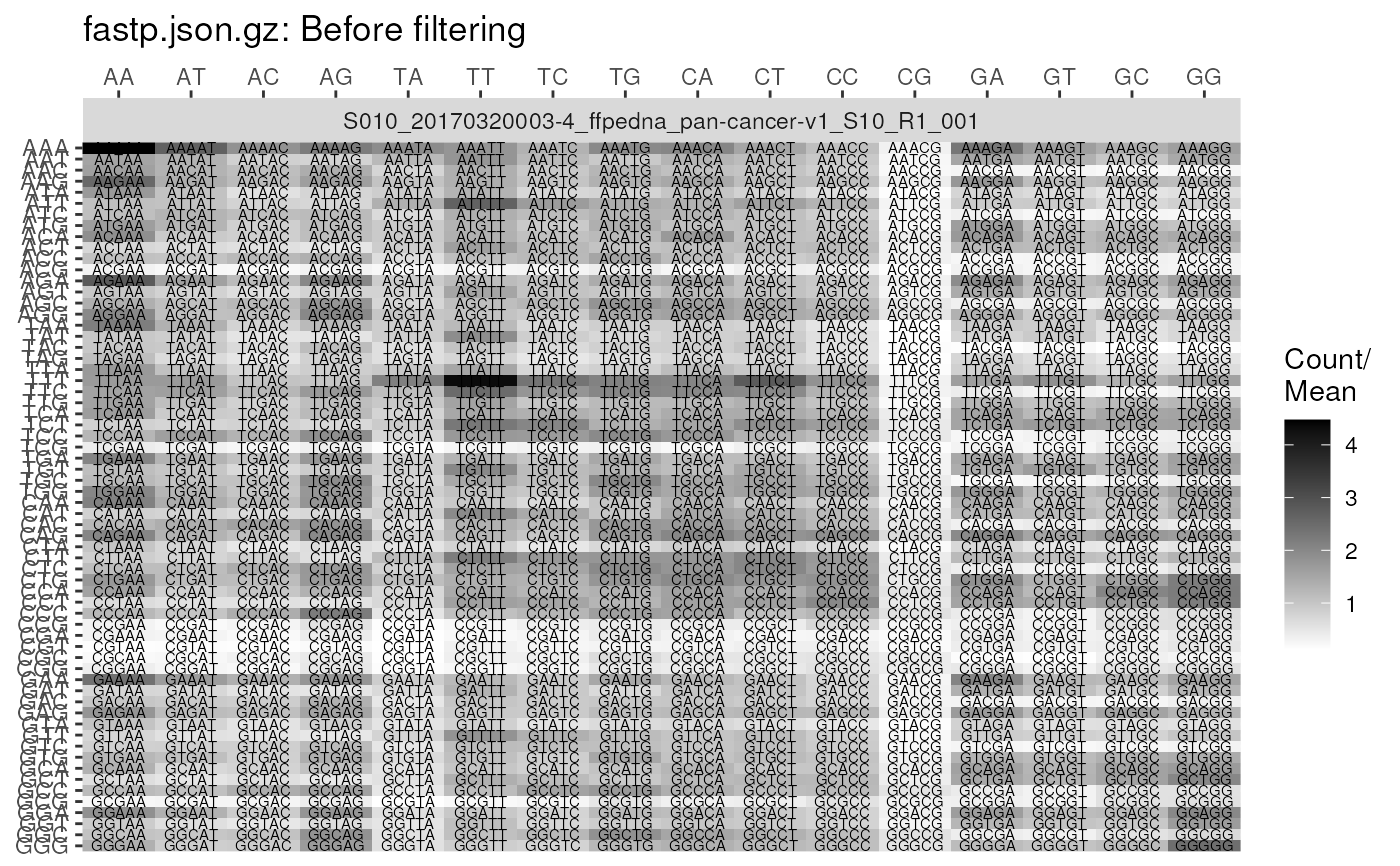
As the Kmer Content module present in FastQC reports is relatively uninformative, and omitted by default in later versions of FastQC, these are rudimentary plots.
Plots for FastqcData objects replicate those contained in a FastQC
report, whilst the heatmap generated from FastqcDataList objects
simply show the location and abundance of over-represented Kmers.
Examples
# Get the files included with the package
packageDir <- system.file("extdata", package = "ngsReports")
fl <- list.files(packageDir, pattern = "fastqc.zip", full.names = TRUE)
# Load the FASTQC data as a FastqcDataList object
fdl <- FastqcDataList(fl)
plotKmers(fdl[[1]])
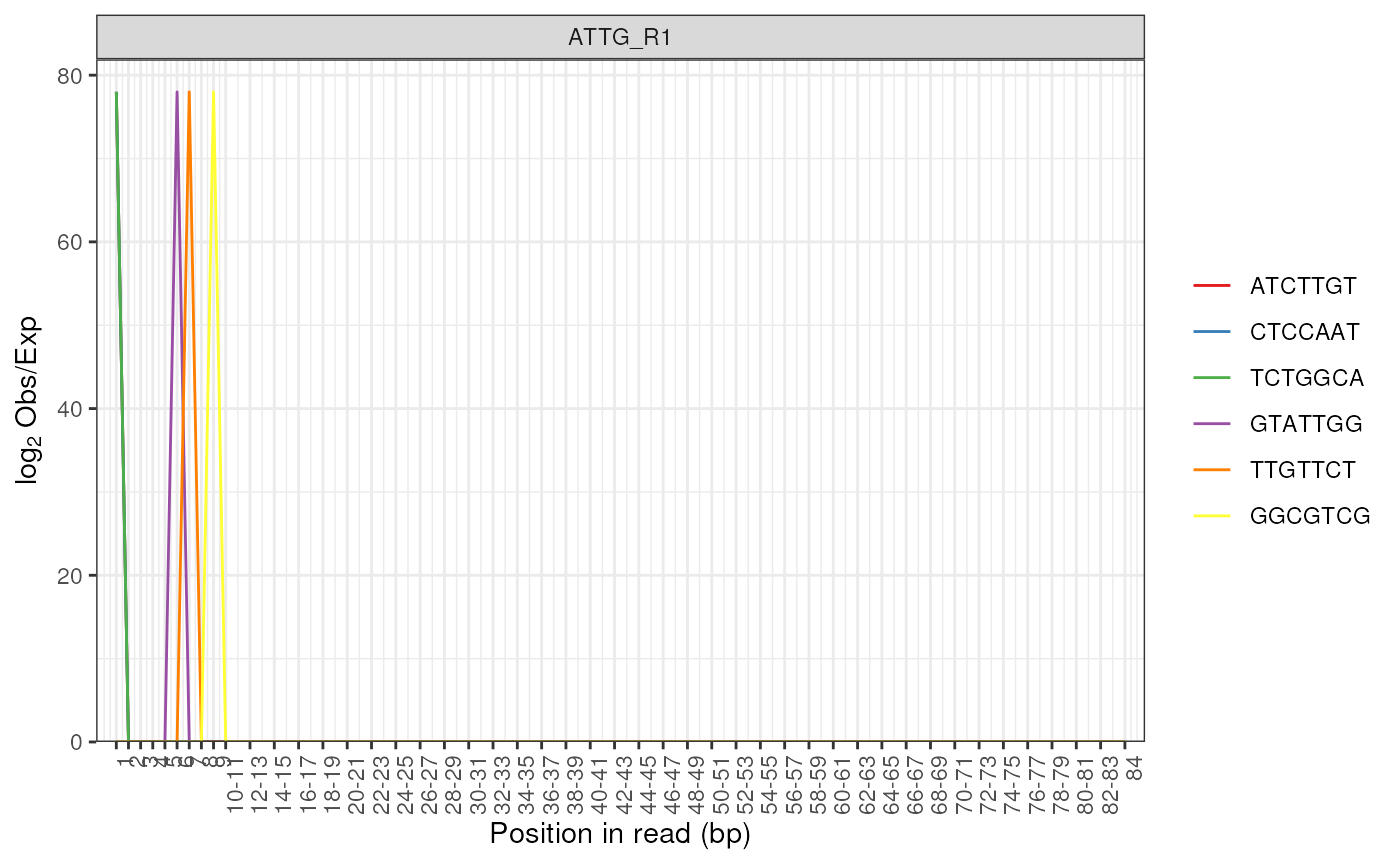 # Use a FastpData object
fl <- system.file("extdata", "fastp.json.gz", package = "ngsReports")
fp <- FastpData(fl)
plotKmers(fp, size = 2)
# Use a FastpData object
fl <- system.file("extdata", "fastp.json.gz", package = "ngsReports")
fp <- FastpData(fl)
plotKmers(fp, size = 2)
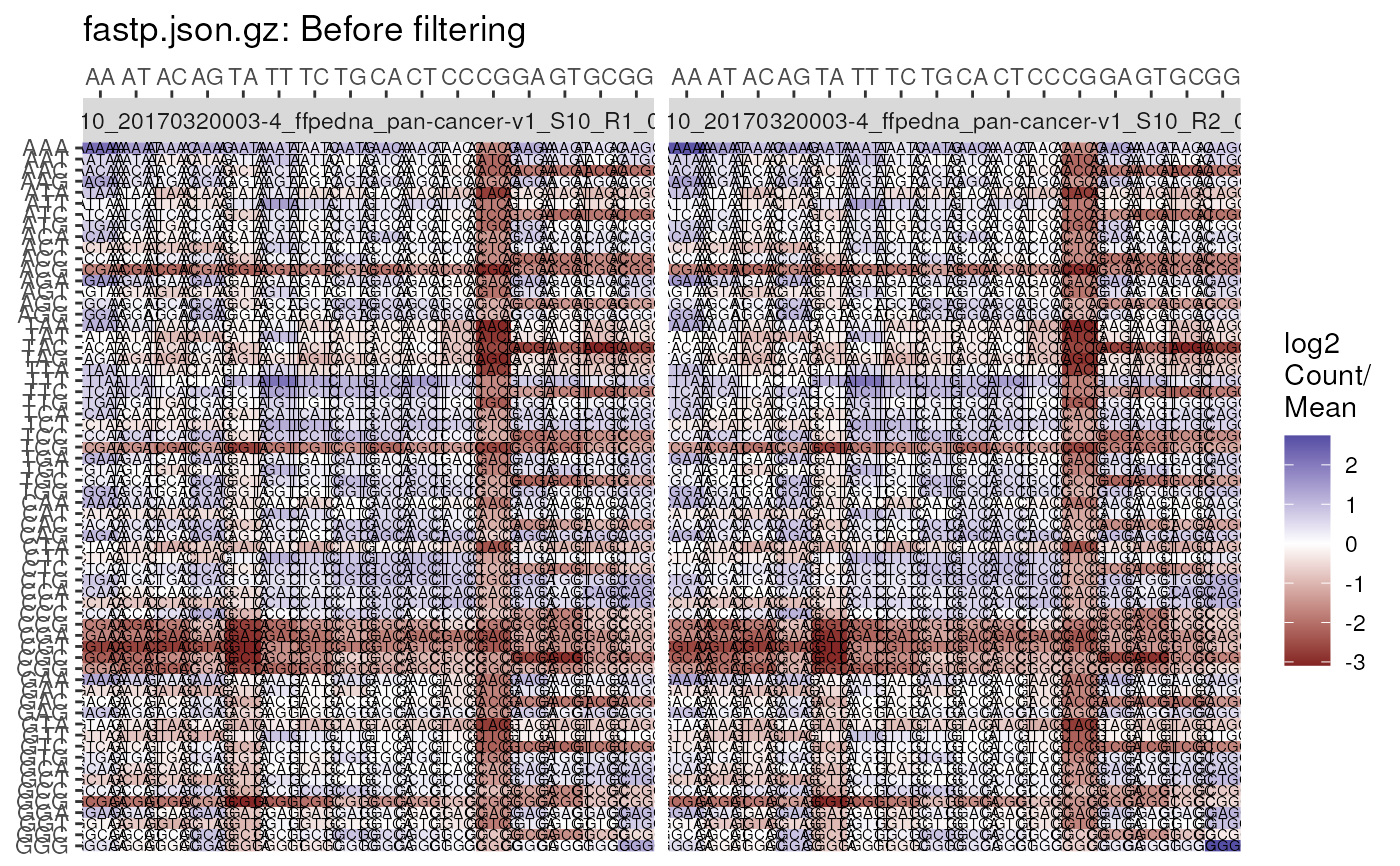 plotKmers(
fp, reads = "read1", size = 2, trans = NULL,
scaleFill = scale_fill_gradient(low = "white", high = "black")
)
plotKmers(
fp, reads = "read1", size = 2, trans = NULL,
scaleFill = scale_fill_gradient(low = "white", high = "black")
)