Plot the Base Qualities for each file as separate plots
plotBaseQuals(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'ANY'
plotBaseQuals(x, usePlotly = FALSE, labels, pattern = ".(fast|fq|bam).*", ...)
# S4 method for class 'FastqcData'
plotBaseQuals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
warn = 25,
fail = 20,
boxWidth = 0.8,
showPwf = TRUE,
plotlyLegend = FALSE,
...
)
# S4 method for class 'FastqcDataList'
plotBaseQuals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
warn = 25,
fail = 20,
showPwf = TRUE,
boxWidth = 0.8,
plotType = c("heatmap", "boxplot"),
plotValue = "Mean",
cluster = FALSE,
dendrogram = FALSE,
nc = 2,
heat_w = 8L,
...
)
# S4 method for class 'FastpData'
plotBaseQuals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
warn = 25,
fail = 20,
showPwf = FALSE,
module = c("Before_filtering", "After_filtering"),
reads = c("read1", "read2"),
readsBy = c("facet", "linetype"),
bases = c("A", "T", "C", "G", "mean"),
scaleColour = NULL,
plotTheme = theme_get(),
plotlyLegend = FALSE,
...
)
# S4 method for class 'FastpDataList'
plotBaseQuals(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
warn = 25,
fail = 20,
showPwf = FALSE,
module = c("Before_filtering", "After_filtering"),
plotType = "heatmap",
plotValue = c("mean", "A", "T", "C", "G"),
scaleFill = NULL,
plotTheme = theme_get(),
cluster = FALSE,
dendrogram = FALSE,
heat_w = 8L,
...
)Arguments
- x
Can be a
FastqcData,FastqcDataListor character vector of file paths- usePlotly
logicalDefaultFALSEwill render using ggplot. IfTRUEplot will be rendered with plotly- labels
An optional named vector of labels for the file names. All filenames must be present in the names.
- pattern
Regex to remove from the end of the Fastp report and Fastq file names
- ...
Used to pass additional attributes to theme() and between methods
- pwfCols
Object of class
PwfCols()to give colours for pass, warning, and fail values in plot- warn, fail
The default values for warn and fail are 30 and 20 respectively (i.e. percentages)
- boxWidth
set the width of boxes when using a boxplot
- showPwf
Include the Pwf status colours
- plotlyLegend
logical(1) Show legend for interactive plots. Only called when drawing line plots
- plotType
characterCan be either"boxplot"or"heatmap"- plotValue
characterType of data to be presented. Can be any of the columns returned by the appropriate call togetModule()- cluster
logicaldefaultFALSE. If set toTRUE, fastqc data will be clustered using hierarchical clustering- dendrogram
logicalredundant ifclusterisFALSEif bothclusteranddendrogramare specified asTRUEthen the dendrogram will be displayed.- nc
numeric. The number of columns to create in the plot layout. Only used if drawing boxplots for multiple files in a FastqcDataList- heat_w
Relative width of any heatmap plot components
- module
Select Before and After filtering when using a FastpDataList
- reads
Create plots for read1, read2 or all when using a FastpDataList
- readsBy
If paired reads are present, separate using either linetype or by facet
- bases
Which bases to include on the plot
- scaleColour
ggplot discrete colour scale, passed to lines
- plotTheme
theme object
- scaleFill
ggplot2 continuous scale. Passed to heatmap cells
Value
A standard ggplot2 object or an interactive plotly object
Details
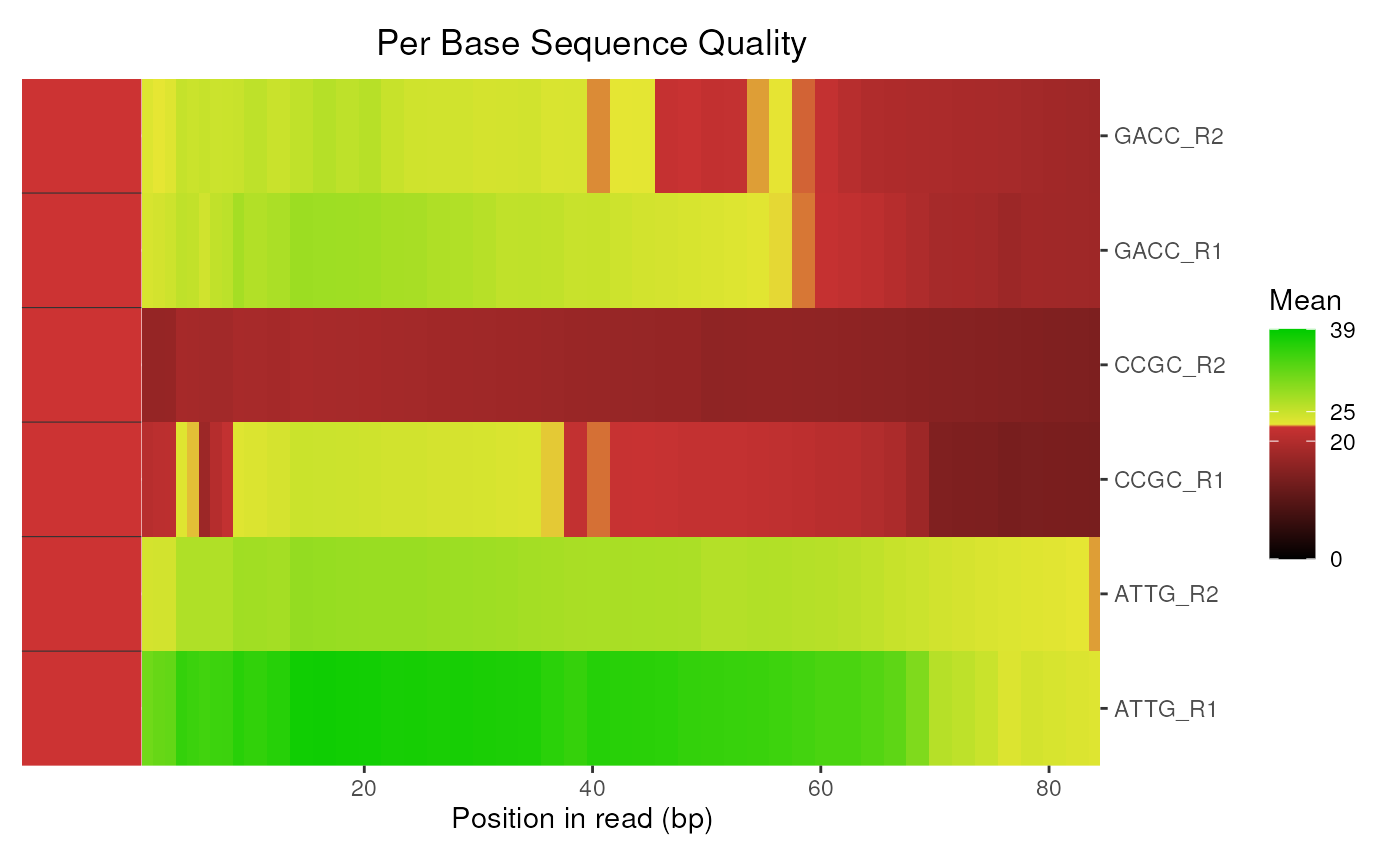
When acting on a FastqcDataList, this defaults to a heatmap
using the mean Per_base_sequence_quality score. A set of plots which
replicate those obtained through a standard FastQC html report can be
obtained by setting plotType = "boxplot", which uses facet_wrap
to provide the layout as a single ggplot object.
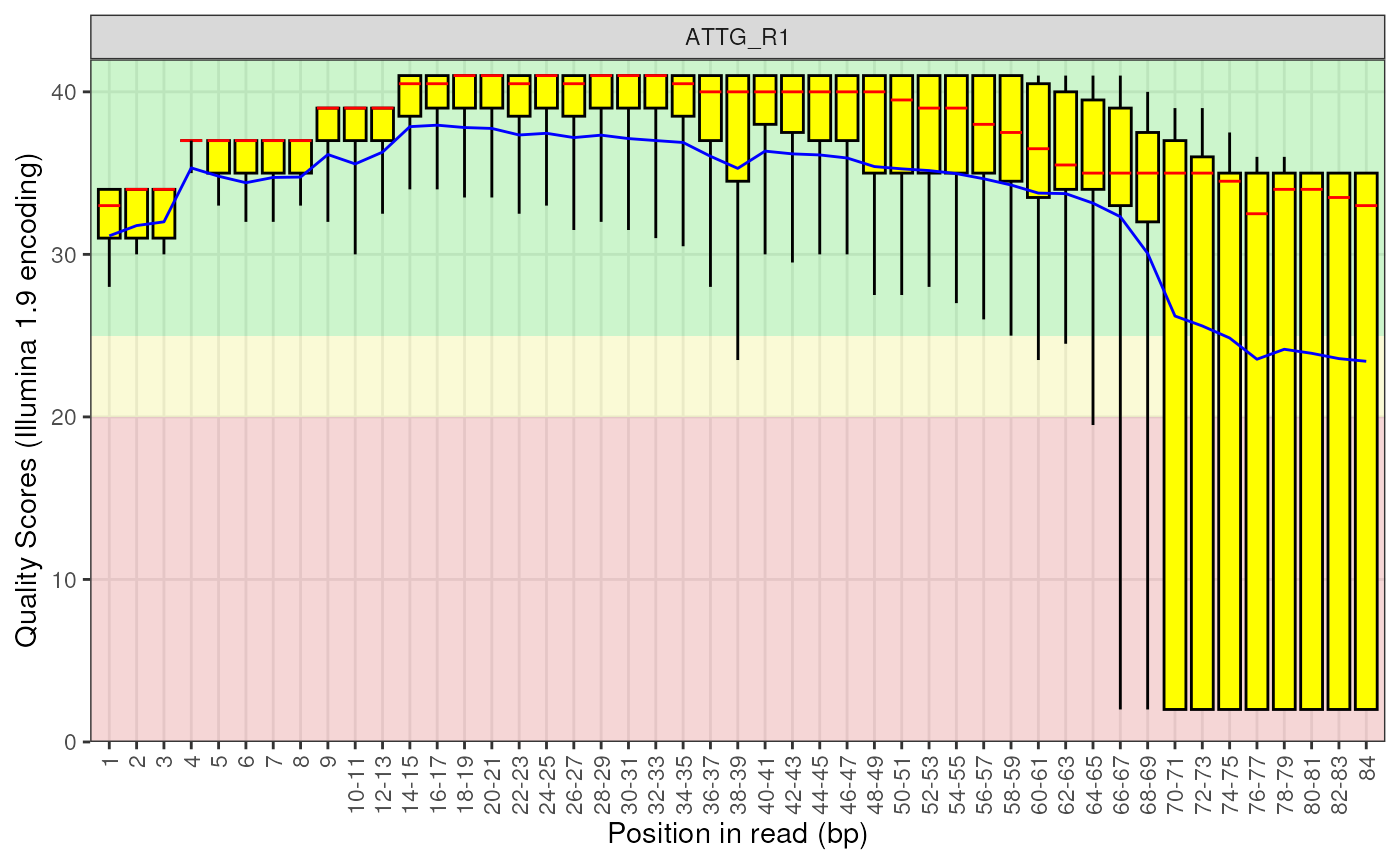
When acting an a FastqcData object, this replicates the
Per base sequence quality plots from FastQC with no faceting.
For large datasets, subsetting by R1 or R2 reads may be helpful.
An interactive plot can be obtained by setting usePlotly = TRUE.
Examples
# Get the files included with the package
packageDir <- system.file("extdata", package = "ngsReports")
fl <- list.files(packageDir, pattern = "fastqc.zip", full.names = TRUE)
# Load the FASTQC data as a FastqcDataList object
fdl <- FastqcDataList(fl)
# The default plot for multiple libraries is a heatmap
plotBaseQuals(fdl)
 # The default plot for a single library is the standard boxplot
plotBaseQuals(fdl[[1]])
# The default plot for a single library is the standard boxplot
plotBaseQuals(fdl[[1]])
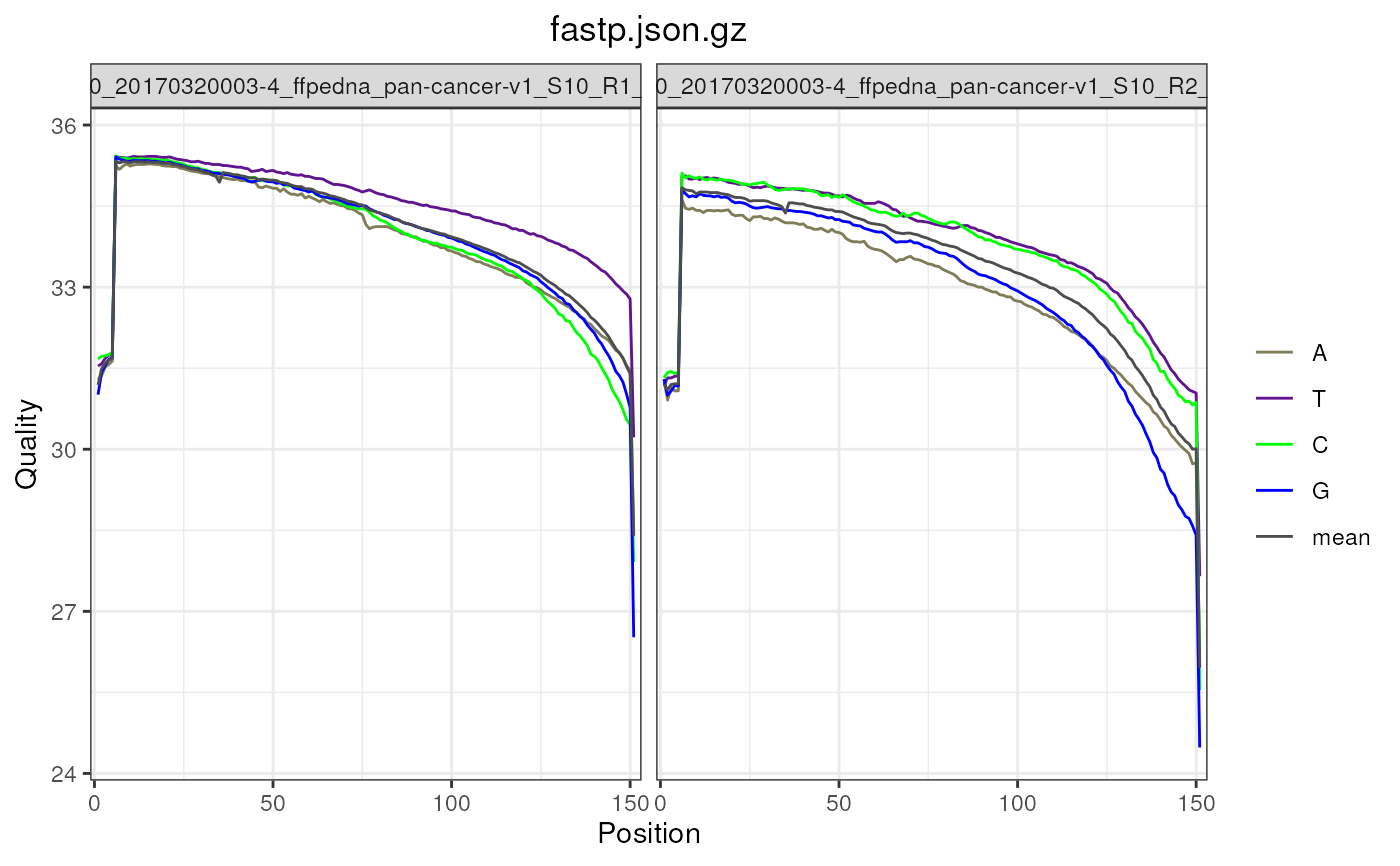
 # FastpData objects have qyalities by base
fp <- FastpData(system.file("extdata/fastp.json.gz", package = "ngsReports"))
plotBaseQuals(
fp, plotTheme = theme(plot.title = element_text(hjust = 0.5))
)
# FastpData objects have qyalities by base
fp <- FastpData(system.file("extdata/fastp.json.gz", package = "ngsReports"))
plotBaseQuals(
fp, plotTheme = theme(plot.title = element_text(hjust = 0.5))
)