Draw an Adapter Content Plot across one or more FASTQC reports
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
...
)
# S4 method for class 'ANY'
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
...
)
# S4 method for class 'FastqcData'
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
showPwf = TRUE,
warn = 5,
fail = 10,
scaleColour = NULL,
plotlyLegend = FALSE,
...
)
# S4 method for class 'FastqcDataList'
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
showPwf = TRUE,
warn = 5,
fail = 10,
plotType = c("heatmap", "line"),
adapterType = "Total",
cluster = FALSE,
dendrogram = FALSE,
heat_w = 8L,
scaleFill = NULL,
scaleColour = NULL,
plotlyLegend = FALSE,
...
)
# S4 method for class 'FastpData'
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
scaleFill = NULL,
plotlyLegend = FALSE,
plotTheme = theme_get(),
...
)
# S4 method for class 'FastpDataList'
plotAdapterContent(
x,
usePlotly = FALSE,
labels,
pattern = ".(fast|fq|bam).*",
pwfCols,
showPwf = FALSE,
warn = 5,
fail = 10,
cluster = FALSE,
dendrogram = FALSE,
scaleFill = NULL,
plotTheme = theme_get(),
heat_w = 8L,
...
)Arguments
- x
Can be a
FastqcData, aFastqcDataListor character vector of file paths- usePlotly
logical. Output as ggplot2 (default) or plotly object.- labels
An optional named vector of labels for the file names. All filenames must be present in the names.
- pattern
regex used to trim the ends of all filenames for plotting
- ...
Used to pass additional attributes to theme() for FastQC objects and geoms for Fastp objects
- pwfCols
Object of class
PwfCols()containing the colours for PASS/WARN/FAIL- showPwf
logical(1) Show PASS/WARN/FAIL status as would be included in a standard FastQC report
- warn, fail
The default values for warn and fail are 5 and 10 respectively (i.e. percentages)
- plotlyLegend
logical(1) Show legend when choosing interactive plots. Ignored for heatmaps
- plotType
character. Can only take the valuesplotType = "heatmap"orplotType = "line"- adapterType
A regular expression matching the adapter(s) to be plotted. To plot all adapters summed, specify
adapterType = "Total". This is the default behaviour.- cluster
logicaldefaultFALSE. If set toTRUE, fastqc data will be clustered using hierarchical clustering- dendrogram
logicalredundant ifclusterisFALSEif bothclusteranddendrogramare specified asTRUEthen the dendrogram will be displayed.- heat_w
Width of the heatmap relative to other plot components
- scaleFill, scaleColour
scale_fill\* and scale_colour_\* objects
- plotTheme
Set theme elements by passing a theme
Value
A standard ggplot2 object, or an interactive plotly object
Details
This extracts the Adapter_Content module from the supplied object and generates a ggplot2 object, with a set of minimal defaults. The output of this function can be further modified using the standard ggplot2 methods.
When x is a single or FastqcData object line plots will always be
drawn for all adapters.
Otherwise, users can select line plots or heatmaps.
When plotting more than one fastqc file, any undetected adapters will not be
shown.
An interactive version of the plot can be made by setting usePlotly
as TRUE
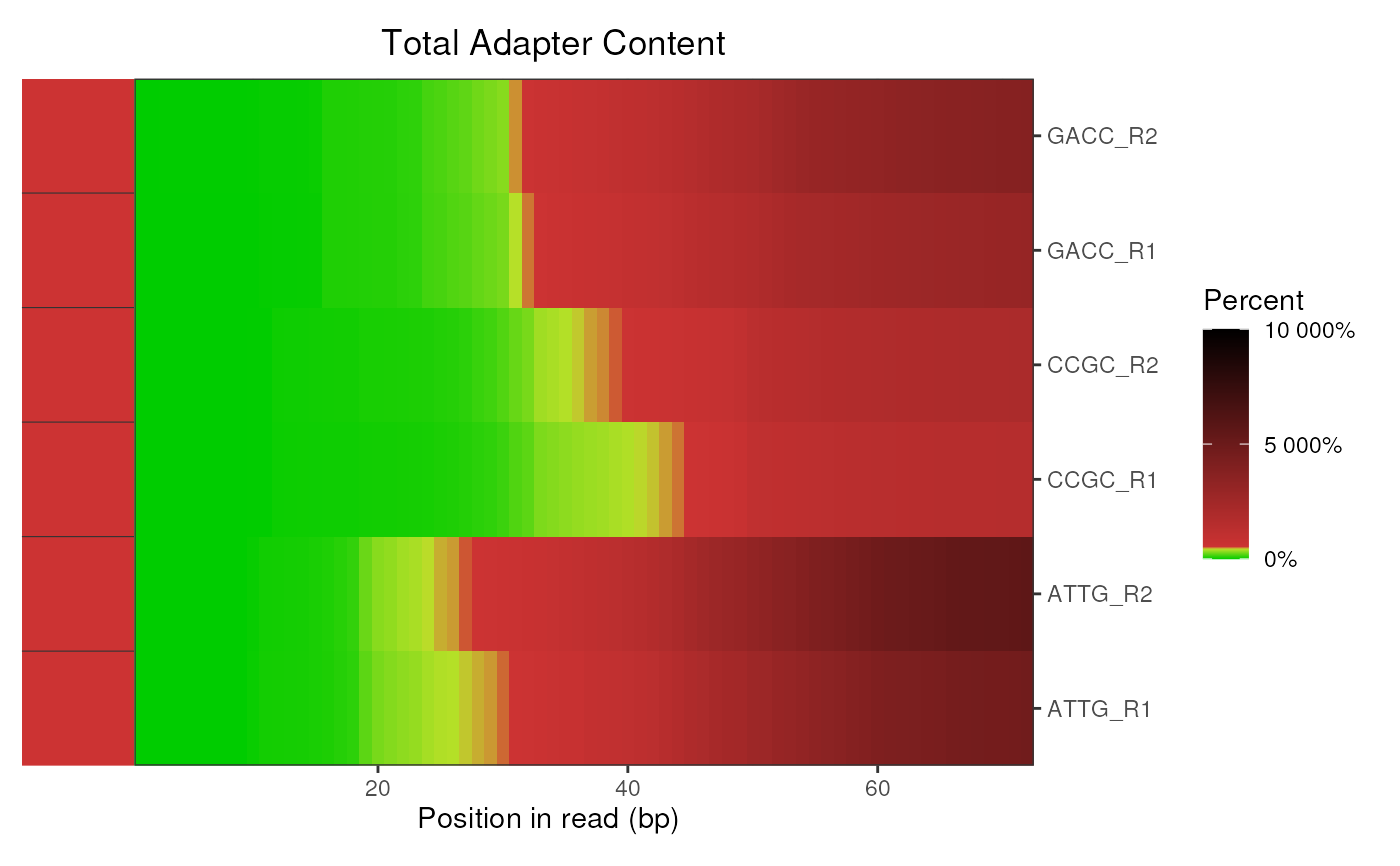
Examples
# Get the files included with the package
packageDir <- system.file("extdata", package = "ngsReports")
fl <- list.files(packageDir, pattern = "fastqc.zip", full.names = TRUE)
# Load the FASTQC data as a FastqcDataList object
fdl <- FastqcDataList(fl)
# The default plot
plotAdapterContent(fdl)
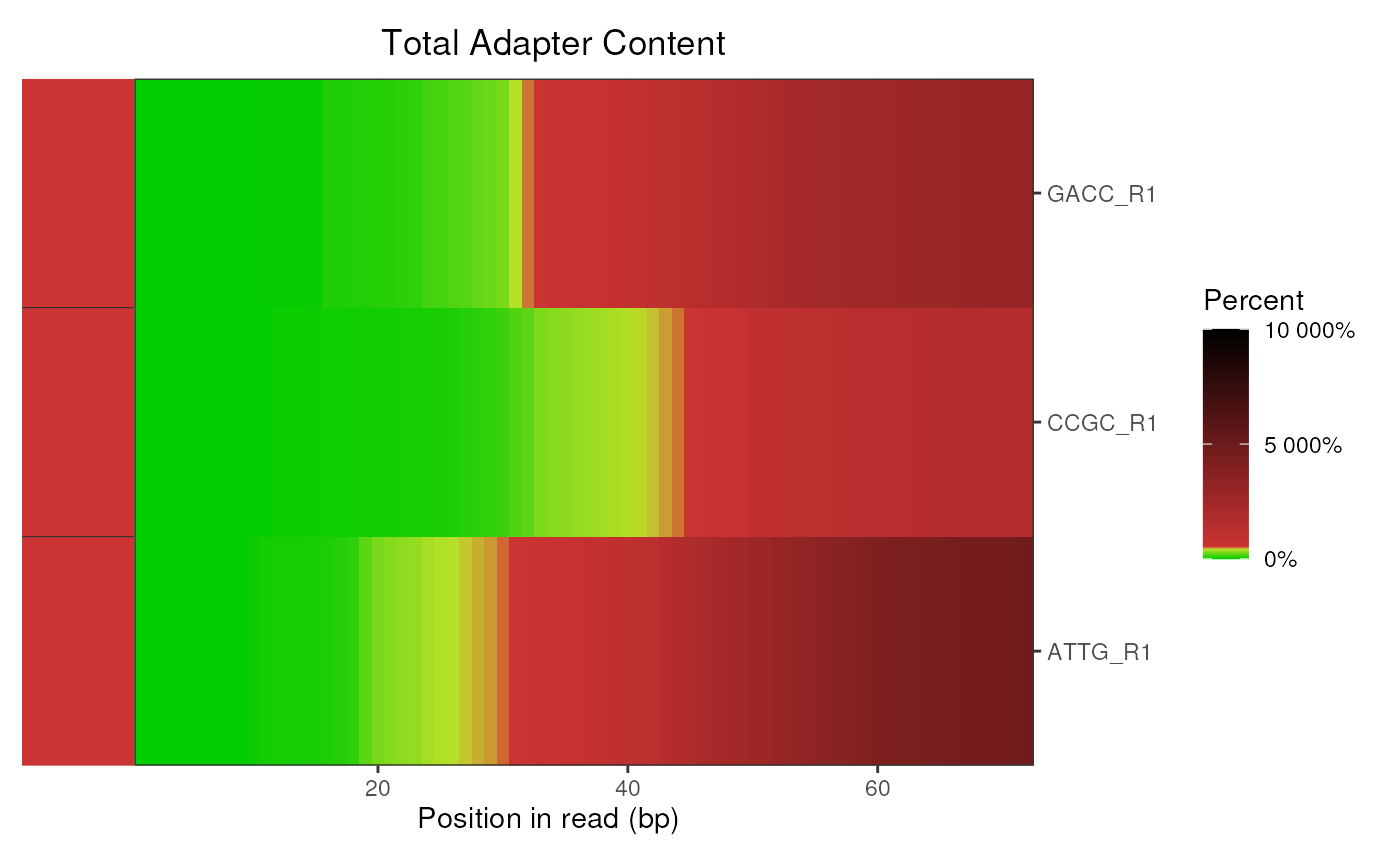
 # Also subset the reads to just the R1 files
r1 <- grepl("R1", fqName(fdl))
plotAdapterContent(fdl[r1])
# Also subset the reads to just the R1 files
r1 <- grepl("R1", fqName(fdl))
plotAdapterContent(fdl[r1])
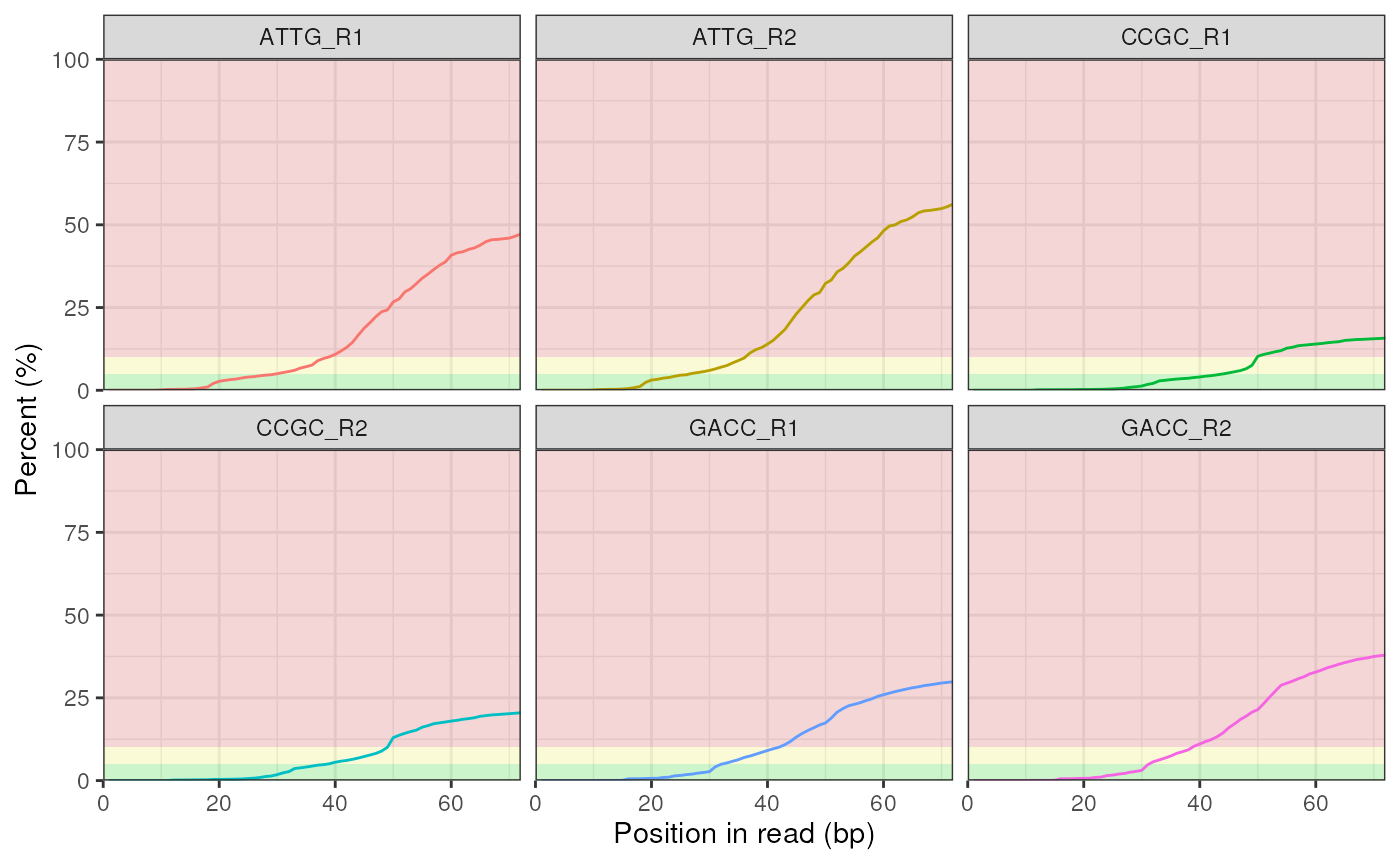
 # Plot just the Universal Adapter
# and change the y-axis using ggplot2::scale_y_continuous
plotAdapterContent(fdl, adapterType ="Illumina_Universal", plotType = "line") +
facet_wrap(~Filename) +
guides(colour = "none")
# Plot just the Universal Adapter
# and change the y-axis using ggplot2::scale_y_continuous
plotAdapterContent(fdl, adapterType ="Illumina_Universal", plotType = "line") +
facet_wrap(~Filename) +
guides(colour = "none")
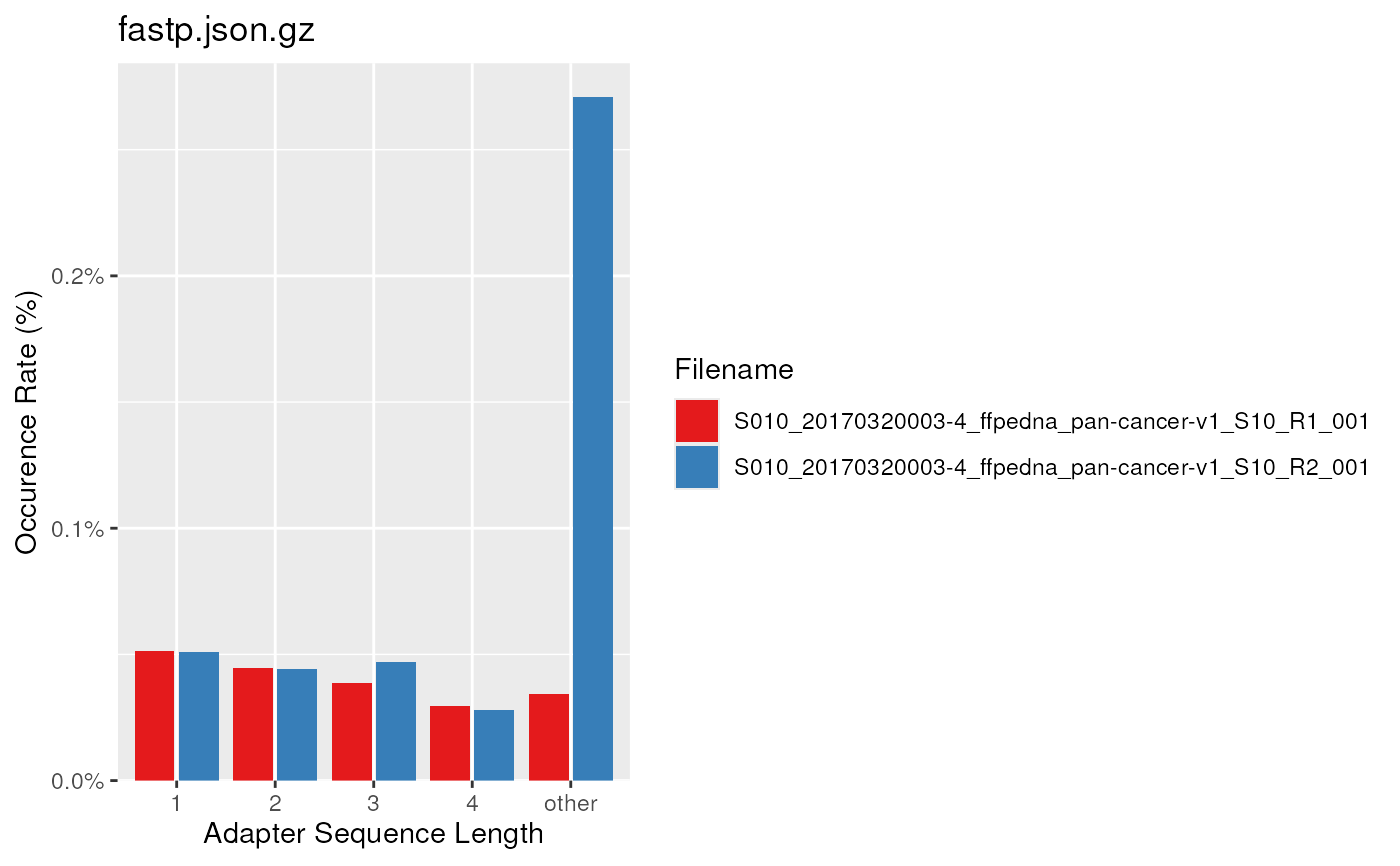
 # For FastpData object, the plots are slightly different
fp <- FastpData(system.file("extdata/fastp.json.gz", package = "ngsReports"))
plotAdapterContent(fp, scaleFill = scale_fill_brewer(palette = "Set1"))
# For FastpData object, the plots are slightly different
fp <- FastpData(system.file("extdata/fastp.json.gz", package = "ngsReports"))
plotAdapterContent(fp, scaleFill = scale_fill_brewer(palette = "Set1"))